databases
It's time for the annual blog about the annual Nucleic Acids Research (NAR) database issue. This is the 24th database issue for NAR and the seventh blog for @finchtalk. Like most years I have no idea what I'm going to write about until I start reading the new issue. Something always inspires me.
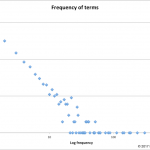
This year's inspiration came from missing data.
In 2017, NAR lists 1662 databases or 23 fewer than last year.
As summarized in the database issue's introduction, Galperin, Fernández-Suarez, and Rigden tell us this year's issue has 152 papers. 54 of those describe new databases, 98…
When finding a female scientists' data turns into an archeological treasure hunt.
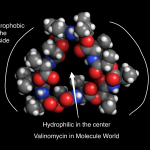
A few months ago, I decided it would be interesting to celebrate various scientific contributions by making images of chemical / molecular structures in the Molecule World iPad app and posting them on Twitter (@MoleculeWorld). Whenever I can, I like to highlight scientific contributions from women on their birthdays. Tomorrow's post will feature Dr. Isabella Karle, an x-ray crystallographer who worked on the Manhattan project and solved structures of interesting molecules like…
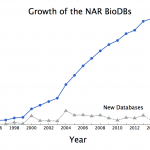
Something interesting happened in 2014. The total number of databases that Nucleic Acids Research (NAR) tracks dropped by three databases!
What happened? Did people quit making databases? No. This year, the "dead" databases (links no longer valid) outnumber the new ones. To celebrate Digital World Biology's release of Molecule World I'll discuss some of the new structure databases below. But first, the numbers.
As summarized in the database issue's introduction, Galperin, Rigden, and Fernández-Suárez tell us this year's issue has 172 papers. 56 of those…
Sometimes when you go digging through the databases, you find unexpected things.
When I was researching the previous posts on insulin structure and insulin evolution, I found something curious indeed.
Human insulin, colored by rainbow. Image from the Molecule World iPad app by Digital World Biology.
I wanted to find out how many different organisms made insulin, so I used a database at the NCBI called Blink. Blink is a database of protein blast search results. Using Blink can save you lots of time because…
On pinene and inhibiting enzymes.
People of a certain age may remember a series of really funny commercials featuring Euell Gibbons and his famous question about whether you've ever eaten a pine tree. "Some parts are edible" said Euell.
Perhaps some parts are, but other pine tree products aren't so nourishing. Crystallography365, aka @Crystal_in_city had a couple of fun blog posts about pinene, a chemical made by pine trees, that also inhibits cytochrome P450 2B6.
I was inspired by their posts and by my experience with Cytochrome P450 to go a little…
By @finchtalk (Todd Smith)
In 2014 and beyond Finchtalk will be contributing to Digitalbio’s blog at this site. We kick off 2014 with Finchtalk’s traditional post on the annual database issue from Nucleic Acids Research (NAR).
Biological data and databases are ever expanding. This year was no exception as the number of databases tracked by NAR grew from 1512 to 1552. In the leadoff introduction [1] the authors summarize this year’s issue and the status of the NAR index. The 21st issue includes 185 articles with 58 new databases and 123 updates. In the 1552 database repository, 193 had their…
Is there a place for citizen scientists in the world of digital biology?
Many of the citizen science projects that I've been reading about have a common structure. There's a University lab at the top, outreach educators in the middle, and a group of citizens out in the field collecting data.
After the data are collected, they end up in a database somewhere and the University researchers analyze them and write papers. At least that's my impression so far.
It seems to me, that with all kinds of databases out there, on-line, there should be plenty of opportunity for both citizens and student…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
Warfarin, a commonly used anti-clotting drug, sold under the brand name of Coumadin, has a been a poster child for the promise of pharmacogenomics and personalized medicine.
The excitement has come from the idea that knowing a patient's genotype, in this case for the VKORC1 and CYP2C9 genes, would allow physicians to tailor the dose of the drug and get patients the correct dose more quickly.
And it seems obvious that a test that would allow doctors to predict your ability to metabolize warfarin, would be a great thing, right?
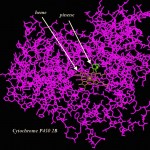
Figure 1. Human Cytochrome P450 Cyp2c9 bound to Warfarin…
Last night, the phone rang at 9:22 pm. I quickly glanced at the caller ID. Hmmm. Why is the Seattle School district calling us at this time of night?
Apparently the swine flu has come to Seattle and the school district thought we should know.
Those messages are helpful if you're a parent, but they don't tell much about the rest of the world.
Health Map is a really wonderful, user-friendly, resource for following the epidemic.
When you get to Health Map , choose Select None to clear the map.
Then select Swine Flu.
You'll see a Google map with markers representing reports. The colors show…
What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
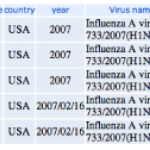
I was pretty impressed to find the swine flu genome sequences, from the cases in California and Texas, already for viewing at the NCBI.
You can get them and work them, too. It's pretty easy. Tomorrow, we'll align sequences and make trees.
Activity 3: Getting the swine flu sequence data
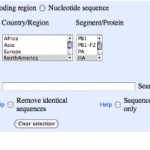
1. Go to the NCBI, find the Influenza Virus Resource page and follow the link to:
04/27/2009: Newest swine influenza A (H1N1) sequences.
2. You'll see a page that looks like this:
Each column heading is a name of a segment of the influenza genome. You can see there are eight of these. Each segment…
I'm a big of learning from data. There are many things we can learn about swine flu and other kinds of flu by using public databases.
In digital biology activity 1, we learned about the kinds of creatures that can get flu. Personally, I'm a little skeptical about the blowfly, but...
Now, you might wonder, what kinds of flu do these different creatures get? Are they all getting H1N1, or do they get different variations? What are H and N anyway?
We can discuss all of these, but for now, lets see what kinds of flu strains infect different kinds of creatures.
Activity 2. What flu infects…
Genome sequences from California and Texas isolates of the H1N1 swine flu are already available for exploration at the NCBI. Let's do a bit of digital biology and see what we can learn.
Activity 1. What kinds of animals get the flu?
For the past few years we've been worrying about avian (bird). Now, we're hearing about swine (pig) flu.
All of this news might you wonder just who gets the flu besides pigs, birds, and humans. We can find out by looking at the data.
Over the past few years, researchers have been sequencing influenza genomes and depositing those genomes in public…
In the first post, I talked about how factual data aren't creative works, and how compiling them into collections doesn't make them creative - at least in the US.
This aspect of data rips away the core "incentive" provided by copyright law to creators: the right to sue people who make copies. It also has a second aspect, which is that the international treaties that govern copyright don't apply. Whatever one may think of those treaties, they do a fair amount to normalize the laws worldwide - a copyright on a Britney Spears tune applies in much the same way in wildly different countries. For…
I got drawn into a debate about copyrights and factual data this week that felt like it merited its own blog post. It was kind of surreal new media debating - I was going back and forth with a smart guy from the UC Berkeley school of information on a friend's Facebook wall for most of a day on the topic. It was definitely a change from the typical FB chatter and in some ways the character count constraints of a wall post were formative to the debate. But some of the questions raised deserved long answers, and the issues involved are complicated and subtle and non-obvious. Hopefully moving the…
I suppose I should have expected this.
I thought it might be fun to see what the databases had to say about turkeys.
Technorati Tags: Thanksgiving,, turkey,, mash-up
So, I queried the NCBI databases, found a taxonomy reference, and started clicking related links to see pictures of the different species.
Why?
Because it would be great to see what the different species of turkies look like and compare them.
Here's a species of wild turkey that I didn't expect to find.
Wild turkey, humph!
Do mosquitoes get the mumps? Part V. A general method for finding interesting things in GenBank
This is the last in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes and a general method for finding other interesting things.
In this last part, I discuss a general method for finding novel things in GenBank and how this kind of project could be a good sort of discovery, inquiry-based project for biology, microbiology, or bioinformatics students.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III.…
Part IV. Assembling the details and making the case for a novel paramyxovirus
This is the fourth in a five part series on an unexpected discovery of a paramyxovirus in a mosquito. In this part, we take a look at all the evidence we can find and try to figure out how a gene from a virus came to be part of the Aedes aegypti genome.
image from the Public Health Library
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a novel mosquito paramyxovirus
V. A…
Part III. Serendipity strikes when we Blink
In which we find an unexpected result when we Blink while looking at the mumps polymerase.
This is the third in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes. And yes, this is where the discovery happens.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
To paraphrase Louis Pasteur,…