genomics
A key concept in science is molecular scale. DNA is a fascinating molecule in this regard.
While we cannot "see" DNA molecules without the aid of advanced technology, a full length DNA molecule can be very long. In human cells, other than sperm and eggs, six billion base pairs of DNA are packaged into 22 pairs of chromosomes, plus two sex chromosomes. Each base pair is 34 angstroms in length (.34 nanometers, or ~0.3 billionths of a meter), so six billion base pairs (all chromosomes laid out head to toe) form a chain that's two-meters long. If we could hang this DNA chain from a hook, it…
Replication fork - http://en.wikipedia.org/wiki/Telomere.
Organisms with linear chromosomes have to solve the problem that DNA replication makes them shorter. This is due to the fact that DNA polymerase can only add bases to the terminal 3'-OH of a DNA chain. The DNA replication initiation complex uses RNA primers to provide the initial 3'-OH and to initiate "lagging" strand synthesis. While one strand can be copied all the way to the end of a chromosome, the other, lagging strand, must be primed at short intervals in order to provide a 3' OH group for DNA polymerase as the…
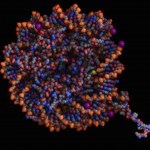
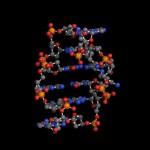
Today (4/25) is national DNA day. Digital World Biology™ is celebrating by sharing some of our favorite structures of DNA. We created these photos with Molecule World™ a new iPad app for viewing molecular structures.
As we are taught in school, the double stranded DNA molecule is a right-handed helix as determined by Watson and Crick using Franklin's x-ray diffraction images [1]. This B-form of DNA has approximately 10 nucleotides per turn of the helix and is the most common form of DNA found in nature.
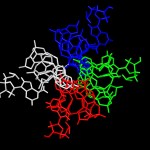
Classic structure with the elements colored.
Classic structure…
In our series on why $1000 genomes cost $2000, I raised the issue that the $1000 genome is a value based on simplistic calculations that do not account for the costs of confirming the results. Next, I discussed how errors are a natural occurrence of the many processing steps required to sequence DNA and why results need to be verified. In this and follow-on posts, I will discuss the four ways (oversampling, technical replicates, biological replicates, and cross-platform replicates) that results can be verified as recommended by Robasky et. al. [1].
The game Telephone teaches us how a…
Previously, I introduced the idea that the $1000 genome has not been achieved because it is defined in simplistic terms that ignore many aspects of data completeness and verification. In that analysis, I cited a recent perspective by Robasky, Lewis, and Church [1] to present concepts related to the need to verify results and the general ways in which this is done. In this and the next few posts I will dig deeper into the elements of sequence data uncertainty and discuss how results are verified.
Agarose gel of DNA stained with ethidium bromide. By Mnolf, wikipedia.
First, we need to…
Getting an accurate genome sequence requires that you collect the data at least twice argue Robasky, Lewis, and Church in their recent opinion piece in Nat. Rev. Genetics [1].
The DNA sequencing world kicked off 2014 with an audacious start. Andrew Pollack ran an article in the New York Times implying that 100,000 genomes will be the new norm in human genome sequencing projects [2]. The article focused on a collaboration between Regeneron and Geisinger Health in which they plan to sequence the exomes (the ~2% of the genome that encodes proteins and some non-coding RNA) of 100,000…
By @finchtalk (Todd Smith)
In 2014 and beyond Finchtalk will be contributing to Digitalbio’s blog at this site. We kick off 2014 with Finchtalk’s traditional post on the annual database issue from Nucleic Acids Research (NAR).
Biological data and databases are ever expanding. This year was no exception as the number of databases tracked by NAR grew from 1512 to 1552. In the leadoff introduction [1] the authors summarize this year’s issue and the status of the NAR index. The 21st issue includes 185 articles with 58 new databases and 123 updates. In the 1552 database repository, 193 had their…
I've heard you have to sing loud if you want to change the world.
Cloning DNA – lyrics by Sandra Porter, sung to the tune of Surfin' USA
C ..................G7..................C
If everybody had a plasmid, across the U.S.A.,
C ..................G7..................C
then everybody'd be cloning, with their DNA
................................F
You'd see them wearing their goggles.
.................... C
and their lab coats, too
.............G7 ...............C
multicolored gloves on, cloning DNA
C…
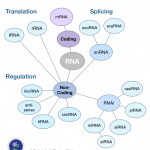
On the Weizmann Wave, researchers have made a discovery surrounding exons—"bits of genetic code that are snipped out of the sequence and spliced together to make the protein instruction list." When a cell needs to make a protein, it pulls exons out of pre-messenger RNA and stitches them together to form messenger RNA. Alternating sequences called introns are left out. By tracking the unused introns, researchers observed that "in some cases, pre-mRNA production shot straight up - to ten times or more than that of the mRNA that followed." They call this "production overshoot," for when "the…
In simple Mendelian genetics, a single change in one gene can produce a large change in mortality. The National Human Genome Research Institute (NHGRI) will be funding genomics studies on Mendelian traits using a similar strategy.
NHGRI will fund a small number of centers, dominant centers you might say, and look for large changes. The sequencing centers that will benefit are the Broad Institute, Washington University, and Baylor College of Medicine. For the next four years, the big three will be dividing $86 million a year according to a press release from NHGRI. I'm not sure what algorithms…
Last week, I applied a little not-so-Respectful Insolence to a movie about a physician and "researcher" named Stanislaw Burzynski, MD, PhD, founder of the Burzynski Clinic and Burzynski Research Institute in Houston. I refer you to my original smackdown for details, but in brief Dr. Burzynski claimed in the 1970s to have made a major breakthrough in cancer therapy through his discovery of anticancer substances in the urine that he dubbed "antineoplastons," which turned out to be mainly modified amino acids and peptides. Since the late 1970s, when he founded his clinic, Dr. Burzynski has been…
For the past few days I've been avidly following Daniel MacArthur's tweets from the Personal Genome Conference at Cold Spring Harbor(@dgmacarthur #cshlpg).
The Personal Genomics tweets aren't just interesting because of the science, they're interesting because MacArthur and others have started to take on the conventional dogma in genetic ethics.
For years, there has been a strong message from the clinical genetics and genetics education community that genetic information is dangerous.
Unlike the other medical tests we're continually urged to get (mammograms, blood pressure readings, sugar…
By way of Matthew Yglesias, we read that,over at National Review Online, Kevin Williamson claims progressives only care about science as a way to wage culture war (yes, coming from movement conservatives, that's rich):
There are lots of good reasons not to wonder what Rick Perry thinks about scientific questions, foremost amongst them that there are probably fewer than 10,000 people in the United States whose views on disputed questions regarding evolution are worth consulting, and they are not politicians; they are scientists. In reality, of course, the progressive types who want to know…
I've blogged before about some of the technical issues surrounding how we can handle the massive increase in the size of genomics datasets. There's also a need to grapple with the analytical aspects of all of these data:
So, from a bacterial perspective, genome sequencing is really cheap and fast--in about a year, I conservatively estimate (very conservatively) that the cost of sequencing a bacterial genome could drop to about $1,500 (currently, commercial companies will do a high-quality draft for around $5,000- $6,000). We are entering an era where the time and money costs won't be…
Update/clarification: I want to clarify something critical. This is not about picking on a researcher or a country. It very well could have happened in the U.S. or anywhere else. I, nor you the reader, have any idea about the internal constraints these groups experience, or what was communicated to government officials. To the extent that data sharing didn't occur due to concerns over publication, this represents an instance where the publication process--and the import attributed to it--affected the need for rapid release. That's the key point, not assigning blame to individuals or…
I recently was in a conversation with a collaborator who isn't in the genomics biz, and said collaborator remarked that there was a lot of online criticism of the quality of the genomic data that has been generated for the E. coli O104:H4 outbreak isolates. I've been following it very closely (not surprised by that, are you?), and I'm not sure what the collaborator was referring to. On some blog, in some comment, there probably is criticism, but these are the intertoobz: that sort of thing happens.
But then it dawned on me that much of what appears to be 'criticism' is probably just a…
Regarding the German outbreak strain of E. coli, the data are fairly clear: it is an enteroaggregative E. coli ('EAEC') which has acquired antibiotic resistance genes and a Shiga-like toxin from an Shiga-toxinogenic E. coli ('STEC'). EAEC are interesting--according to the European Food Safety Authority:
EAEC have been implicated as a cause of persistent diarrhea in children and acquired immunodeficiency syndrome-associated diarrhea, as well as acute diarrhea in travelers. Not all EAEC strains have been shown to cause diarrhea in humans. The EAEC are a heterogeneous group of bacteria that…
After Friday's post, I've held off on writing much about the German E. coli outbreak, often referred to by its serotype, O104:H4, or as HUSEC041 (HUS stands for hemolytic uremic syndrome). Having had the weekend to digest some of the ongoing analysis and news reports, here are some additional thoughts:
1) The multilocus sequence type (MLST) of this outbreak is definitely ST678. This means this outbreak strain is related to an older strain found in 2001 that caused disease. A new, improved assembly released by BGI yields a perfect match to ST678. In addition, there is independent…
...in Europe. I'll get to that in a moment. You've probably heard of the E. coli outbreak sweeping through Germany and now other European countries that has caused over one thousand cases of hemolytic uremic syndrome ('HUS'). What's odd is that the initial reports are calling this a novel hybrid or some new strain of E. coli.
BGI has done some sequencing using Ion Torrent of one of these isolates, and Nick Loman assembled the data. Without getting too technical, the genome is actually in about 3,000 pieces, but with those data (and thanks to Nick for assembling them and releasing them) I…
The Wall Street Journal reports an estimate of the economic impact of the Human Genome Project (italics mine):
Of the $3.8 billion federal funding for the human-genome project, $2.8 billion originated at the U.S. National Institutes of Health, and the rest at the Department of Energy.
That $3.8 billion, along with subsequent capital provided by the government and the private sector, generated a total return of roughly $49 billion in direct and indirect federal tax revenues over the last two decades or so. (The $3.8 billion is worth about $5.6 billion in constant 2010 dollars.)
Over the same…