What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
NCBI influenza resources
I was pretty impressed to find the swine flu genome sequences, from the cases in California and Texas, already for viewing at the NCBI.
You can get them and work them, too. It's pretty easy. Tomorrow, we'll align sequences and make trees.
Activity 3: Getting the swine flu sequence data
1. Go to the NCBI, find the Influenza Virus Resource page and follow the link to:
04/27/2009: Newest swine influenza A (H1N1) sequences.
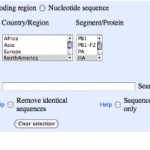
2. You'll see a page that looks like this:
Each column heading is a name of a segment of the influenza genome. You can see there are eight of these. Each segment…