Molecules of sucrose tore apart in their bellies letting glucose course free in their veins.
Molecules of sucrose tore apart in their bellies letting glucose course free in their veins.
Luckily for us, a system evolved long ago to capture that glucose and minimize it's potential for damage. Removing sugar from the blood and sequestering it in liver, fat, and muscle cells, minimizes the harm that might result if sugars were free to bind to proteins and our bodies were stressed by trying to flush excess glucose out of our system.
This holiday season, we give thanks for insulin and the biotech companies like Genentech that cloned the human gene and began to produce this protein in genetically modified microbes.
![]() Today, insulin is one of the most successful biotech drugs of all time and a few companies use DNA technology to manufacture it in yeast and bacteria. Diabetics worldwide are grateful for this technology since it allows them to manage their disease and minimize the possibility of blindness and damage to kidneys and nerves.
Today, insulin is one of the most successful biotech drugs of all time and a few companies use DNA technology to manufacture it in yeast and bacteria. Diabetics worldwide are grateful for this technology since it allows them to manage their disease and minimize the possibility of blindness and damage to kidneys and nerves.
 Glucose
Glucose
What is insulin?
Insulin is a hormone made of protein that our pancreas makes when the sugar levels in our blood get too high. Cells in the pancreas secrete insulin into the blood stream where it binds to other cells with insulin receptors on their surface. When insulin binds to those receptors, it stimulates the production of glucose transport proteins that can vacuum up glucose and move it into cells.
Interestingly, the small size and medical importance of insulin made it’s structure one of the first to be solved. Dorothy Hodgkin's laboratory solved the structure of insulin by X-ray crystallography in 1969 (1).
What can we learn about insulin by looking at the structure?
Dr. David Goodsell, wrote a really nice story about the insulin structure in 2001 (3). At that time, only smaller structures from insulin had been solved. Today, we have new models that represent an additional form that insulin takes in the body. This makes it worthwhile to reexamine the kinds of information we can learn from studying these structures. Links within and at the end of this article will let you download the structures and explore them yourselves in Molecule World or Cn3D. All the images in this post were created with a pre-release version of the Molecule World™ iPad app (version 2).
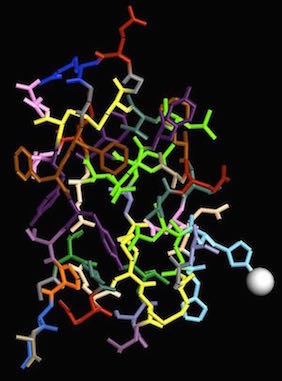 Pig insulin, PDB ID 4INS Pig insulin, PDB ID 4INS
|
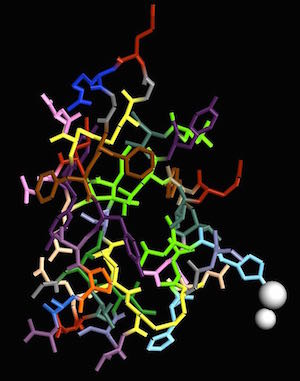 Human insulin, PDB ID 1TRZ Human insulin, PDB ID 1TRZ
|
Before the insulin gene was cloned, and the protein made available through biotechnology, drug companies obtained insulin by isolating it from pigs. Pig insulin works in humans because the structure is very similar. In the images above, I used residue coloring to show each amino acid with a different color and emphasize the similarity between pig and human structures. of all the amino acids in the insulin protein, only one differs between the pig and human forms.
Both pig (4INS)(3) and human (1TRZ) (4) insulin contain two chains of amino acid residues and one atom of zinc. The insulin gene only codes for one protein chain but that original protein gets cut into two pieces by a protease. Disulfide bonds between cysteine residues hold the two chains together.
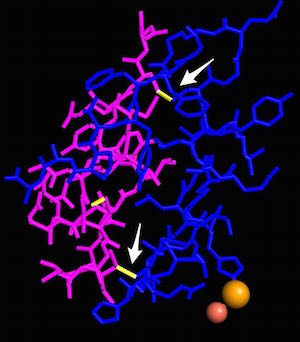 Disulfide bonds between chains A and B in human insulin, PDB ID 1TRZ
Disulfide bonds between chains A and B in human insulin, PDB ID 1TRZ
Leucines in the center of the monomer also contribute to the final three dimensional structure.
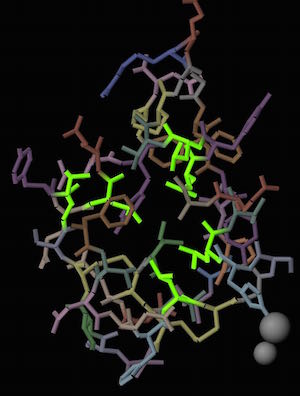 Leucines in the center of the human insulin monomer are highlighted in green.
Leucines in the center of the human insulin monomer are highlighted in green.
Taking a single chain and coloring it by rainbow, shows how the protein folds into the three dimensional shape.
 Coloring a single protein chain by rainbow shows that protein folding places one end of the amino acid sequence very close to the other.
Coloring a single protein chain by rainbow shows that protein folding places one end of the amino acid sequence very close to the other.
These monomeric units represent only one of the forms that insulin proteins can take. The insulin that diabetics take is usually in a hexametric form, composed of six insulin monomers with two atoms of zinc (5).
 Human insulin, 4F51, element coloring. Human insulin, 4F51, element coloring.
|
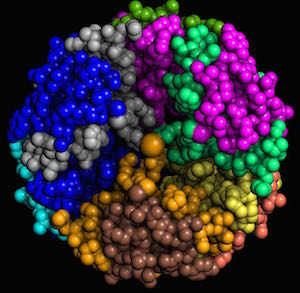 Human insulin, 4F51. Each chain is shown in a different color. Human insulin, 4F51. Each chain is shown in a different color.
|
If I use charge coloring to identify where the positive and negatively charged amino acids are located, I can see where histidines hold the zinc in place.
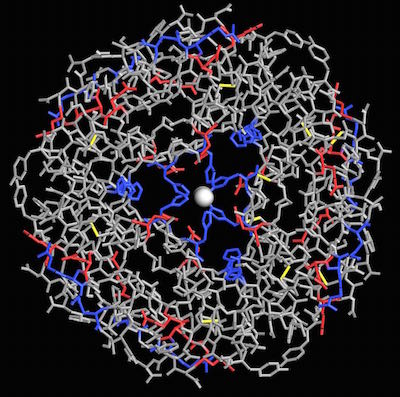 Histidines (blue) in human insulin (4F51) bind to zinc in the center of the hexamer.
Histidines (blue) in human insulin (4F51) bind to zinc in the center of the hexamer.
To summarize our results, we can learn many things about insulin by examining molecular models from different forms of the insulin structure. By viewing and comparing structures, we found:
1. Pig and human insulin are very similar. (Can you spot the one amino acid difference in the images above?)
2. Disulfide bonds hold the two parts of an insulin monomer together.
3. Leucines are present in the center of the insulin monomer.
4. The A chain folds, bringing one end of the amino acid chain near the other.
5. The form of insulin produced by drug companies has six monomers with zinc in the center.
6. Zinc is a part of the insulin monomer and hexamer and is held there through a bond to histidine.
Images: All the images in this article were made from a soon-to-be released version of the Molecule World™ iPad app (version 2) (Digital World Biology). The structures were obtained from the NCBI's Molecular Modeling Database. The structures used were: 4INS (pig monomer), 1TRZ (human monomer), 4F51 (human hexamer).
References:
1. BLUNDELL, T., CUTFIELD, J., CUTFIELD, S., DODSON, E., DODSON, G., HODGKIN, D., MERCOLA, D., & VIJAYAN, M. (1971). Atomic Positions in Rhombohedral 2-Zinc Insulin Crystals Nature, 231 (5304), 506-511 DOI: 10.1038/231506a0
2. Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD, Sakabe K, Sakabe N, Vijayan NM (1988). The Structure of 2ZN PIG Insulin Crystals at 1.5 Angstroms Resolution Philos.Trans.R.Soc.London, Ser.B PMID: 2905485
3. Goodsell, D. (2001). Insulin RCSB Protein Data Bank DOI: 10.2210/rcsb_pdb/mom_2001_2
5. Ciszak E, & Smith GD (1994). Crystallographic evidence for dual coordination around zinc in the T3R3 human insulin hexamer. Biochemistry, 33 (6), 1512-7 PMID: 8312271
5. Fávero-Retto, M., Palmieri, L., Souza, T., Almeida, F., & Lima, L. (2013). Structural meta-analysis of regular human insulin in pharmaceutical formulations European Journal of Pharmaceutics and Biopharmaceutics, 85 (3), 1112-1121 DOI: 10.1016/j.ejpb.2013.05.005