bioinformatics
What do you call a biologist who uses bioinformatics tools to do research, but doesn't program?
You don't know?
Neither does anyone else.
The names we use
People who practice biology are known by many names, so many, that the number of names almost reflects the diversity of biology itself.
Sometimes we describe biologists by the subject they study. Thus, we have biologists from anatomists to zoologists, and everything in between: addiction researchers, chronobiologists, epidemiologists, immunologists, microbiologists, neuroscientists, pharmacologists, physiologists, plant biologists,…
I often get questions about bioinformatics, bioinformatics jobs and career paths. Most of the questions reflect a general sense of confusion between creating bioinformatics resources and using them. Bioinformatics is unique in this sense. No one confuses writing a package like Photoshop with being a photographer, yet for some odd reason, people seem to expect this of biologists. In the same respect, even the programmers and database administrators who work in bioinformatics, are unfairly assumed to have had graduate level training in biology.
In many ways, it's easiest to understand…
For many years, I've been perfectly content to work with small numbers of things. Working with one gene or one protein is great. Even small groups of genes are okay. I'm fine with alternatively spliced genes with multiple transcripts, or multiple polymorphisms, or genes in multi-gene families, or groups of genes in operons.
But I never trusted microarrays.
First, there were all the articles questioning the ability to reproduce microarray data. For many years, people reported difficulties in reproducing experiments, especially if they used chips from different manufacturers or different…
Last spring, I gave my first hands-on workshop in working with Next Generation Sequencing data at the Eighth Annual UT-ORNL-KBRIN Bioinformatics Summit at Fall Creek Falls State Park in Tennessee. The proceedings from that conference are now on-line at BMC Bioinformatics and it's fun to look back and reflect on all that I learned at the conference and all that's happened since.
Figure 1. Fall Creek Falls State Park, TN
When the conference took place, Geospiza had only just released new versions of GeneSifter Analysis Edition that could do gene expression analysis with Next Gen data.
Who…
For those of you who may have been wondering where I've been, these past few weeks have seen me grading final projects, writing a chapter on analyzing Next Gen DNA sequencing data for the Current Protocols series, and flying back and forth between Seattle and various meetings elsewhere in the U.S. It will probably take years of bike commuting to make up for my carbon credits, but most meetings I attend don't have viable alternatives in venues like Second Life or World of Warcraft. Anyway, as I sit writing on an airplane, I think I could revise the title for Dr. Seuss' famous book to "Oh the…
Should any data, not just genomic data, be held hostage by the grant award process?
Hunh? Let me back up...
By way of ScienceBlogling Daniel MacArthur, I came across this excellent post by David Dooling about, among other things, how different genome centers, based on size, have different release policies (seriously, read his post). Dooling writes (boldface mine):
The more interesting question is: why aren't all data and research released rapidly and freely available? Since the Bermuda Principles were agreed to in 1996, all genome sequencing centers have submitted their data, from raw…
I realize that the typical format for blogging is to find something that pisses you off and then rant about it, but I actually like the recent workshop report by NHGRI, "The Future of DNA Sequencing at the National Human Genome Research Institute." (pdf file) While I'll have more to say about the report overall, I liked the section about the Human Microbiome Project (the goal of the HMP is to use sequencing technologies to understand how the microbes that live on us and in us affect health and disease).
I was happy to see that NHGRI still thinks that it has a role in funding the HMP. It's…
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done…
We'll have a blast, I promise! But there's one little thing we need to discuss first...
I want to explain why I'm going to use nucleotide sequences for the blast search. (I used protein the other day). It's not just because someone told me too, there is a solid rational reason for this.
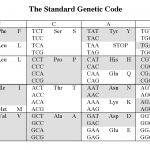
The reason is the redundancy in the genetic code.
Okay, that probably didn't make any sense to those of you who didn't already know the answer. Here it is.
The picture above shows the human genetic code (there are at least 16 variations on this, but that's another story). Each middle cell in the…
We had a great discussion in the comments yesterday after I published my NJ trees from some of the flu sequences.
If I list all the wonderful pieces of advice that readers shared, I wouldn't have any time to do the searches, but there are a few that I want to mention before getting down to work and posting my BLAST results.
Here were some of the great suggestions and pieces of advice;
1. Do a BLAST search. Right! I can't believe I didn't do that first thing, I think the trees I got surprised me so much all sense flew out of my brain.
2. Show us the multiple alignments. Okay. I'll…
What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
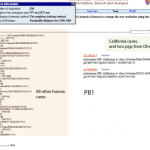
This afternoon, I was working on educational activities and suddenly realized that the H1N1 strain that caused the California outbreak might be the same strain that caused an outbreak in 2007 at an Ohio country fair.
UPDATE: I'm not so certain anymore that the strains are the same. I'm doing some work with nucleic acid sequences to look further at similarity.
Here's the data.
Once I realized that the genome sequences from the H1N1 swine flu were in the NCBI's virus genome resources database, I had to take a look.
And, like eating potato chips, making phylogenetic trees is a little bit…
I was pretty impressed to find the swine flu genome sequences, from the cases in California and Texas, already for viewing at the NCBI.
You can get them and work them, too. It's pretty easy. Tomorrow, we'll align sequences and make trees.
Activity 3: Getting the swine flu sequence data
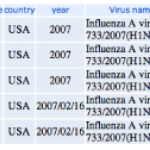
1. Go to the NCBI, find the Influenza Virus Resource page and follow the link to:
04/27/2009: Newest swine influenza A (H1N1) sequences.
2. You'll see a page that looks like this:
Each column heading is a name of a segment of the influenza genome. You can see there are eight of these. Each segment…
I'm a big of learning from data. There are many things we can learn about swine flu and other kinds of flu by using public databases.
In digital biology activity 1, we learned about the kinds of creatures that can get flu. Personally, I'm a little skeptical about the blowfly, but...
Now, you might wonder, what kinds of flu do these different creatures get? Are they all getting H1N1, or do they get different variations? What are H and N anyway?
We can discuss all of these, but for now, lets see what kinds of flu strains infect different kinds of creatures.
Activity 2. What flu infects…
Genome sequences from California and Texas isolates of the H1N1 swine flu are already available for exploration at the NCBI. Let's do a bit of digital biology and see what we can learn.
Activity 1. What kinds of animals get the flu?
For the past few years we've been worrying about avian (bird). Now, we're hearing about swine (pig) flu.
All of this news might you wonder just who gets the flu besides pigs, birds, and humans. We can find out by looking at the data.
Over the past few years, researchers have been sequencing influenza genomes and depositing those genomes in public…
A couple of years ago, I answered a reader's question about the cost of genome sequencing. One of my readers had asked why the cost of sequencing a human genome was so high. At that time, I used some of the prices advertised by core labs on the web and the reported coverage to estimate the cost of sequencing Craig Venter's genome. As you can imagine, the cost of sequencing has dropped quite a bit since then.
In 2007, Genome Technology reported the cost of sequencing Venter's genome was $70 million. Watson's genome at only $2 million, was a bargain.
Why was Watson's genome so cheap?
Even…
I don't usually publish press releases, but I'm making an exception for this one, since your's truly is one of the Co-PI's. If you're a teacher within commuting distance of Seattle, the schedule and sign up information is here.
NSF AWARDS $1.3 MILLION TO NWABR FOR BIOINFORMATICS EDUCATION
Innovative Technology Experiences for Students and Teachers (ITEST) brings the understanding of how biology and information technology interact to teachers and their students
Seattle, WA - March 26, 2009 - The Northwest Association for Biomedical Research (NWABR) has been awarded a $1.3 million dollar…
In which we identify unknown human proteins.
Yesterday, I wrote about using the BLOSUM 62 matrix to calculate a score for matches between two proteins. Those scores give us a good start on understanding how blastp determines whether two sequences are matching by chance or because they're more likely to be related. But that's not all there is to calculating a blast score, and there's at least one other statistic to consider as well, the E value.
It all comes down to biochemistry
The BLOSUM 62 matrix is based on the substitutions that really do or do not happen in real protein sequences. I…
In which we search for Elvis, using blastp, and find out how old we would have to be to see Elvis in a Las Vegas club.
Introduction
Once you're acquainted with proteins, amino acids, and the kinds of bonds that hold proteins together, we can talk about using this information to evaluate the similarity between protein sequences. We can easily imagine that if two protein sequences are identical, then those proteins would have the same kind of activity. But what about proteins that are similar in some regions, and not others, or proteins that only share some of the same amino acids in similar…
John Hawks recounts a recent conversation about bioinformatics:
I was talking with a scientist last week who is in charge of a
massive dataset. He told me he had heard complaints from many of his
biologist friends that today's students are trained to be computer
scientists, not biologists. Why, he asked, would we want to do that
when the amount of data we handle is so trivial?
Now, you have to understand, to this person a dataset of 1000 whole genomes is
trivial. He said, don't these students understand that in a few years
all the software they wrote to handle these data will be obsolete?…